
The field of Parkinson's disease (PD) remains dynamic, with scientific and industry activities contributing to an accumulation of knowledge and approaches. David Weinstein, CEO of Lunai Bioworks (NASDAQ: LNAI), a phenomics-driven drug discovery company, notes that these efforts may help open new avenues for more tailored approaches and potentially improve options available to patients in the future. "I believe the work that matters most is work that listens to the data, clinicians, and patients, and then lets those voices shape where we test ideas. We have now identified subgroups of Parkinson's, and we believe our platform will generate therapeutic options for these subgroups," he says.
To frame this perspective, Weinstein emphasizes a long‑standing clinical understanding. According to his view, current therapeutic options in Parkinson's disease are primarily directed toward alleviating symptoms rather than altering the underlying course of the condition. Weinstein also notes that investigational agents aiming for disease modification have not yet produced definitive evidence of halting progression in randomized studies. In his assessment, no single intervention comprehensively addresses the diverse range of challenges faced by patients.
"We describe PD through observable symptoms, for example, depression-like mood shifts, slowing of movements, instability of balance, and a range of other impairments, which, I believe, provide a necessary but incomplete map of what is happening biologically," Weinstein states. Lunai approaches this incomplete map by treating PD as a set of overlapping subtypes rather than a single, uniform disease. Weinstein adds, "That conceptual pivot invites a different type of inquiry that looks for patterns within patient heterogeneity and then aligns interventions with those patterns."
Lunai's model begins with a structured effort to bring disparate patient signals into a single analytic frame. The company's phenograph is designed to gather clinical observations, symptomatic descriptors, and other patient-level attributes and to organize them into patterns that can be explored computationally.
The Augusta platform is introduced in that fabric: a set of tools that includes its own curated associations between genes and disease (called its Phenograph) and a machine learning process that can find coherent patient subgroups and biological signatures. "Neither component is presented as a black box or a one-size-fits-all solution," Weinstein emphasizes. Rather, they're framed as instruments for prioritization, ways to help focus research on the spots where evidence may come together most clearly.
When cohorts begin to take shape, a different set of activities follows. Lunai explains a pathway from cluster identification to mechanistic hypothesis generation to targeted experimental validation. The company positions this as a precision-minded discovery. Once a subgroup is characterized by a repeatable set of features, researchers can construct a working model of that subtype, propose plausible biological mechanisms, and seek interventions that address those mechanisms. Their newest work leverages proteomics to identify molecular signatures that correspond with these fine patient groups. These signatures can be used as potential biomarkers or targets for therapeutic development through Lunai's platform.
"If you can describe the patient you aim to help with more clarity, the interventions you design can be clearer in purpose," Weinstein notes. Essentially, the shift is away from broadly applied symptomatic treatments and toward therapies that are matched to a more specific biological context.
That translation phase is supported by an in vivo system Lunai has developed around zebrafish. According to Weinstein, these animals provide a living context in which genetic edits and candidate compounds can be observed at scale and with temporal resolution. Lunai points out that because early development and organ formation can be filmed, machine vision systems can capture behavioral, structural, and physiological shifts as they unfold in real time.
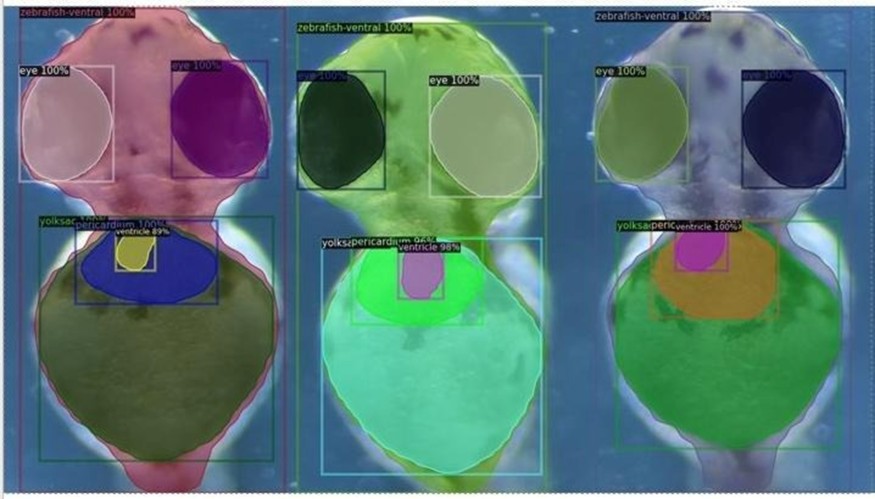
"Our approach is to introduce targeted genetic or molecular perturbations that mimic a specific subtype, observe how symptoms manifest, then apply candidate agents and measure whether the intervention alters the trajectory," Weinstein states. Lunai characterizes this as a rapid, iterative loop designed to reduce guesswork and reveal toxicity signals early. Weinstein believes that this capacity may help translate computational predictions into greater experimental confidence.
Underneath these operational layers is an emphasis on data stewardship. "We've made efforts to reduce variability introduced by different measurement instruments and to standardize inputs before machine learning is applied. The intent is to help increase the likelihood that discovered patterns reflect biology rather than artifacts of how data were gathered," Weinstein states. This practical step is framed as a prerequisite for interpretability and for building models that can be meaningfully aligned with clinical observation.
Overall, these elements form a continuous workflow that aims to bridge observation and intervention. The approach doesn't promise immediate cures or sweeping claims about outcomes. Instead, it seeks to make the pathway from patient signal to therapeutic hypothesis more explicit and more testable. "Our tools are an invitation to biology to tell its story in more detail," Weinstein says, "and to researchers to build responses that are specific rather than speculative."
© 2026 ScienceTimes.com All rights reserved. Do not reproduce without permission. The window to the world of Science Times.












